Efficient Coding
Principles and Practices for Performant Code
Agenda
- Measuring Performance: Time and profile your code
- Common Performance Tweaks: Easy wins for faster code
- Loops vs. Vectorisation vs. …: Choose the right approach
- Optimising Loops: When you should use them
- Beyond the Basics: Tools for further optimisation
Measuring Performance
Timing
- How long does it take?
- Can compare approaches?
- When will your code finish running when you scale it up?
🐍 Timing
🦜 Timing
system.time()for quick one-off timing- {bench} for parameterised comparisons
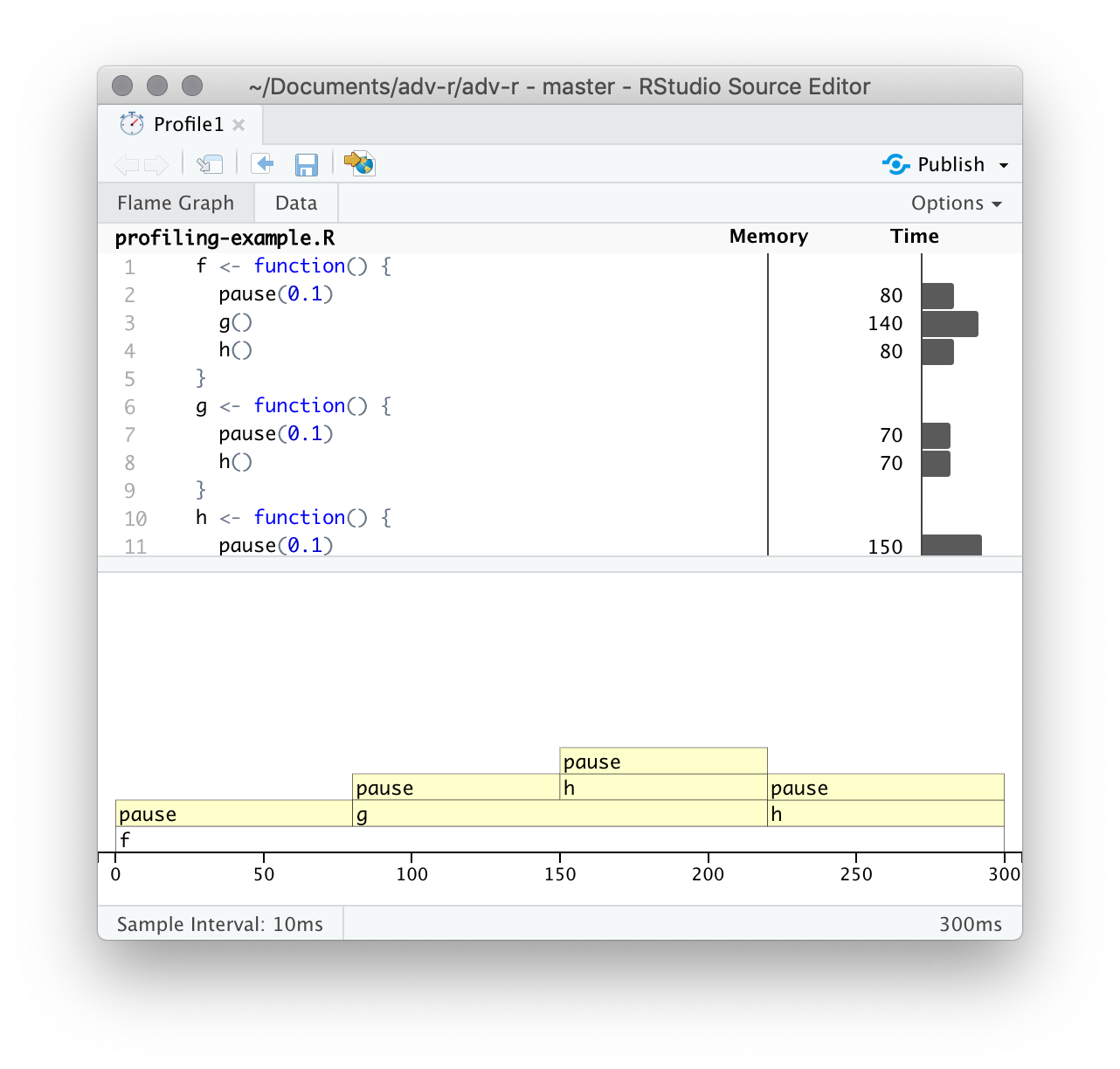
Profiling
“We should forget about small efficiencies, say about 97% of the time: premature optimization is the root of all evil.” - Donald Knuth
- Identify the slowest functions
🐍 Profiling
Code
from random import uniform
from pyinstrument import Profiler
# Monte Carlo method estimating pi through simulation and geometric probability
def hits(point):
return abs(point) <= 1
def point():
return complex(uniform(0, 1), uniform(0, 1))
def estimate_pi(n):
return 4 * sum(hits(point()) for _ in range(n)) / n
with Profiler(interval=0.1) as profiler:
estimate_pi(n=10_000_000)
profiler.print()
# profiler.open_in_browser()🦜 Profiling
Performance Tweaks
Object growth
It starts small… 🥎
![]() {fig-alt=“A garage full of bikes and sports equipment .fragment width=”70%}
{fig-alt=“A garage full of bikes and sports equipment .fragment width=”70%}
🐍 Pre-allocating Arrays
Code
from timeit import timeit
import numpy as np
size = 100_000
def growing_list():
result = []
for i in range(size):
result.append(i**2)
return result
def preallocated_array():
result = np.zeros(size, dtype=int)
for i in range(size):
result[i] = i**2
return result
t1 = timeit(growing_list, number=100)
t2 = timeit(preallocated_array, number=100)
print(f"Growing list: {t1:.6f}s\nPre-allocated: {t2:.6f}s")
print(f"Speedup: {t2/t1:.1f}x faster")🦜 Pre-allocating Arrays
| n | 1 | 2 |
|---|---|---|
| 10^5 | 0.208 | 0.024 |
| 10^6 | 25.50 | 0.220 |
| 10^7 | 3827 | 2.212 |
Appropriate Data Structures
- NumPy array faster than a Python list
- Set is much faster than list* but only keeps unique elements
- Just doing numeric calculations? Can you use a matrix?
🐍 Appropriate Data Structures
| Data Structure | Mutability | Use Cases | Performance |
|---|---|---|---|
| List | Mutable | Ordered collections | Moderate |
| Tuple | Immutable | Fixed collections | Fast |
| Dictionary | Mutable | KV pairs, fast lookups | Fast |
| Set | Mutable | Unique collections | Fast |
| NumPy Array | Mutable | Numerical data, math. ops | Very Fast |
Loops vs. Vectorisation vs. …
🐍 Loops
Code
from timeit import timeit
size = 64
def standard_loop():
result = []
for i in range(size):
result.append(2**i)
return result
def list_comprehension():
return [2**i for i in range(size)]
t1 = timeit(standard_loop, number=100)
t2 = timeit(list_comprehension, number=100)
print(f"Standard loop: {t1:.6f}s\nList comprehension: {t2:.6f}s")
print(f"Speedup: {t1/t2:.1f}x faster")🐍 Vectorisation with NumPy
Code
from timeit import timeit
import numpy as np
size = 100_000
def python_way():
return [i**2 for i in range(size)]
def numpy_way():
return np.arange(size)**2 # Uses C implementation
t1 = timeit(python_way, number=100)
t2 = timeit(numpy_way, number=100)
print(f"Python: {t1:.6f}s\nNumPy: {t2:.6f}s")
print(f"Speedup: {t1/t2:.1f}x faster")🐍 Vectorisation with Pandas
Code
import pandas as pd
import numpy as np
from timeit import timeit
df = pd.DataFrame({"value": np.random.rand(10_000)})
def apply_method():
return df["value"].apply(lambda x: x**2)
def vector_method():
return df["value"]**2
# Compare execution times
t1 = timeit(apply_method, number=100)
t2 = timeit(vector_method, number=100)
print(f"apply: {t1:.6f}s\nvectorised: {t2:.6f}s")
print(f"Speedup: {t1/t2:.1f}x faster")🦜 Vectorisation
🐍 Functional Programming
🐍 Generators
When to Use Each Approach
| Approach | Best For | Example Use Case |
|---|---|---|
| Loops | Complex logic, small data | Custom algorithms |
| Vectorisation | Numerical operations | Data science, NumPy |
| Functional | Data transformations | Pipelines, filter/map/reduce |
| List Comprehensions | Simple transformations | Filter elements |
| Generators | Large dataset processing | Read large files line by line |
Loop Optimisation
Optimisation Techniques
- Define anything you can outside the loop
- Consider locally assigning common functions
- I/O slows loops
- Look out for
printorplot - Use flag for “chatty” / “quiet”
- Proper logging instead of printing
🐍 Optimisation Techniques
Code
from timeit import timeit
import math
from random import randint
size = 300_000
data = [2**randint(0, 64) for _ in range(size)]
def regular_loop():
result = 0
for i in range(len(data)):
x = data[i]
result += math.sqrt(x) + math.sin(x) + math.cos(x)
return result
def optimised_loop():
result = 0
n = len(data)
sqrt, sin, cos = math.sqrt, math.sin, math.cos
for i in range(n):
x = data[i]
result += sqrt(x) + sin(x) + cos(x)
return result
t1 = timeit(regular_loop, number=100)
t2 = timeit(optimised_loop, number=100)
print(f"Regular: {t1:.6f}s\nOptimised: {t2:.6f}s")
print(f"Speedup: {t1/t2:.1f}x faster")🦜 Optimisation Techniques
Beyond the Basics
Rewrite in C++
🐍 Just-In-Time Compilation (1)
Code
from numba import jit
import numpy as np
from timeit import timeit
def slow_func(x):
total = 0
for i in range(len(x)):
total += np.sin(x[i]) * np.cos(x[i])
return total
@jit(nopython=True)
def fast_func(x):
total = 0
for i in range(len(x)):
total += np.sin(x[i]) * np.cos(x[i])
return total
x = np.random.random(10_000)
t1 = timeit(lambda: slow_func(x), number=100)
t2 = timeit(lambda: fast_func(x), number=100)
print(f"Python: {t1:.6f}s\nNumba: {t2:.6f}s")
print(f"Speedup: {t1/t2:.1f}x faster")🐍 Just-In-Time Compilation (2)
What is JIT?
JIT (Just-In-Time) compilation translates code into machine code at runtime to improve execution speed. This approach can improve performance by optimising the execution of frequently run code segments.
Key Benefits: - Can provide 10-100x speed-ups for numerical code - Works especially well with NumPy operations - Requires minimal code changes (just add decorators)
Parallel processing
Pros
Larger datasets
Speed
It’s easy to set up
Cons
Debugging is harder
Can be OS specific
Many statistical techniques are fundamentally serial
Can be slower than serial execution due to overheads
Best Practices Summary
- Measure first - profile before optimising
- Use appropriate data structures for the task
- Vectorise numerical operations when possible
- Avoid premature optimisation - readable code first
- Know when to use loops, comprehensions, or functional styles
📚 Resources
Appendix
Appendix: Cython (Basics)
Pure Python version (slow.py):
Cython version (fast.pyx):
Result: Typically 20-100x faster performance
Appendix: Cython (Best Practices)
Key techniques for maximum performance:
# 1. Declare types for all variables
cdef:
int i, n = 10_000 # Integer variables
double x = 0.5 # Floating point
int* ptr # C pointer
# 2. Use typed memoryviews for arrays (faster than NumPy)
def process(double[:] arr): # Works with any array-like object
cdef int i
for i in range(arr.shape[0]):
arr[i] = arr[i] * 2 # Direct memory access
# 3. Move Python operations outside loops
cdef double total = 0
py_func = some_python_function # Store reference outside loop
for i in range(n):
total += c_only_operations(i)
# 4. Use nogil for parallel execution with OpenMP
cpdef process_parallel(double[:] data) nogil: # No Python GIL
# Can now use OpenMP for parallelismAppendix: Cython (Compiling)
Option 1: Using setuptools (recommended for projects)
Option 2: Quick development with pyximport
Option 3: Direct compilation
 {fig-alt=“A garage full of bikes and sports equipment .fragment width=”70%}
{fig-alt=“A garage full of bikes and sports equipment .fragment width=”70%}